This week in lab is going to be the last one I ever do. This is such a surreal feeling. It seems like last week I was so excited to be a part of the program that I could barely process what had happened. Now its two years later and I'm transferring to the University of Arizona in a few short weeks. Life travels at such break neck speeds, making it nearly impossible to really comprehend what is happening while it happens.
How do you wrap up two years? I want to say deep meaningful things but all I can think is "where am I going to find a lab of such welcoming, interesting people ever again?" I'm going to miss you all so much! I can really be myself with you guys.
Wednesday, May 4, 2016
Estrella Mountain Poster 2nd place winner
I guess I forgot to blog last week. Here is what my poster looked like by the time I was done with it.
I won second place!
Wednesday, April 13, 2016
The Slightly-Better Managed Week
Hey all! This week in lab has barely started. I'm blogging early so I can spend more time procrastinating on my research paper rough draft and poster. Whew I cant believe its already that time of year again!
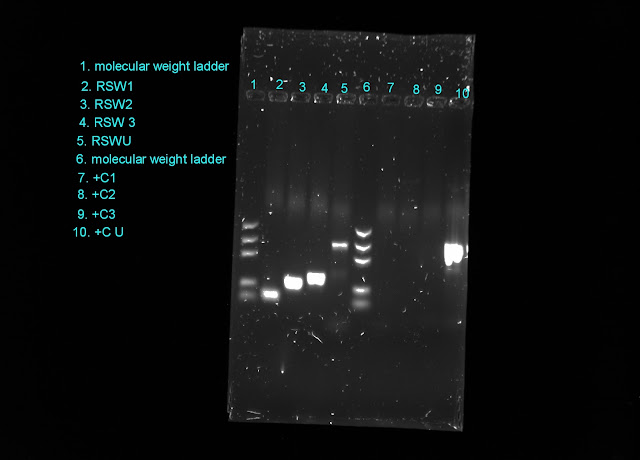
Last week I ran one more PCR on my DNA samples to see if I could get them to work. This time Matt fiddled with the protocol a little bit, making the temperature changes' duration longer. We were hoping to get some better results this time.
Instead the results we got were strange and conflicting. The universal primer Matt gave me amplified all four of my samples but the widow primers I used worked less then 50% of the time. The strangest result was that none of the widow spider primers amplified the known widow spider web DNA (called +C in this experiment). Unfortunately the sample size I'm using is too small and does not allow for any real statistical analysis. I'm not really sure where to go from here.
Last week I ran one more PCR on my DNA samples to see if I could get them to work. This time Matt fiddled with the protocol a little bit, making the temperature changes' duration longer. We were hoping to get some better results this time.
Instead the results we got were strange and conflicting. The universal primer Matt gave me amplified all four of my samples but the widow primers I used worked less then 50% of the time. The strangest result was that none of the widow spider primers amplified the known widow spider web DNA (called +C in this experiment). Unfortunately the sample size I'm using is too small and does not allow for any real statistical analysis. I'm not really sure where to go from here.
Thursday, April 7, 2016
The Mismanaged Week
This week has been long and unproductive, or at least it feels that way. I feel that I don't manage my time as well as I could and end up paying for it at the end of the semester. I always feel like I have plenty of time to do things, put stuff off til the last minuet, and am always rushing to get things done.
So this Tuesday, instead of going to lab, I went to my mom's house and filed my taxes. I owe but no matter. So now I'm rushing at the end of the week to get some lab hours in.
After another inconclusive PCR I've decided to change some of the steps in the PCR protocol and see if that helps. If I don't get clear banding this time I'll have to run another DNA extraction because I'm out of DNA from one of my samples.
Here are my newest gels, see how the banding is not clear except HWUB and FSEUB. The A designation is the first PCR I did run through the thermocycler one more time to see if that increased visible bands. The B is another PCR I ran to see if the mixing of the sample was the problem.
So this Tuesday, instead of going to lab, I went to my mom's house and filed my taxes. I owe but no matter. So now I'm rushing at the end of the week to get some lab hours in.
After another inconclusive PCR I've decided to change some of the steps in the PCR protocol and see if that helps. If I don't get clear banding this time I'll have to run another DNA extraction because I'm out of DNA from one of my samples.
Here are my newest gels, see how the banding is not clear except HWUB and FSEUB. The A designation is the first PCR I did run through the thermocycler one more time to see if that increased visible bands. The B is another PCR I ran to see if the mixing of the sample was the problem.
Thursday, March 31, 2016
PCR Gels 1
This week in lab I took my final PCR product and ran it on a gel to see if the Polymerase Chain Reaction cut my DNA into the correct size of small fragments. The type of gel used for PCR products is exactly the same as the one used for genomic DNA, as well as the dye used to make DNA glow and the machine used to visualize the DNA. So in theory, if I had strong bands of genomic DNA after extraction, and I use the same gels and dye, as long as the PCR worked correctly, I should get pretty strong, clear bands of PCR product.
Unfortunately when I ran my PCR gels they did not come out as beautiful as my genomic DNA bands did. The PCR product was very difficult to visualize using the UV machine, and even where bands could be seen they were not of the right length for the primers I choose. I feel this might have been because I didn't mix my samples enough before putting them into the PCR machine. I am running another trial on my PCR to see if that was the case. If I still have weak bands I will have to trouble shoot the problem again.
Unfortunately when I ran my PCR gels they did not come out as beautiful as my genomic DNA bands did. The PCR product was very difficult to visualize using the UV machine, and even where bands could be seen they were not of the right length for the primers I choose. I feel this might have been because I didn't mix my samples enough before putting them into the PCR machine. I am running another trial on my PCR to see if that was the case. If I still have weak bands I will have to trouble shoot the problem again.
Friday, March 25, 2016
Dilution Fail
This week in lab I ran a Polyermerase Chain Reaction for the first time. I wanted to be able to have the PCR done on Thursday so I could run a gel on Friday but, alas, I did not account for the time it would take to prepare my chemicals and was not able to complete the experiment. All day Thursday I diluted my stock solutions of primers and my DNA samples to have consistent concentrations of each in my tiny test tubes.
To be honest I'm a little annoyed that I didn't know to get that stuff diluted on Tuesday when I had so little to do in the lab. From now on I need to take more initiative and find out exactly what I need to do myself, not rely on the lab instructor when they say I didn't have any prep work.
Anyway today I ran my PCR. One basically mixed together very small quantities of clear liquids into baby sized PCR tubes and then puts them into a machine that does all the work. The PCR machine cycles between various temperatures causing the clear liquids to do different steps of a reaction depending on the temperature. Its all very technical. I'm not sure if mine will work because I didn't mix the tiny little drops when I added them to the tube.
Tiny Tiny Little PCR Tubes:

Tiny Tiny Little Clear Drops:

To be honest I'm a little annoyed that I didn't know to get that stuff diluted on Tuesday when I had so little to do in the lab. From now on I need to take more initiative and find out exactly what I need to do myself, not rely on the lab instructor when they say I didn't have any prep work.
Anyway today I ran my PCR. One basically mixed together very small quantities of clear liquids into baby sized PCR tubes and then puts them into a machine that does all the work. The PCR machine cycles between various temperatures causing the clear liquids to do different steps of a reaction depending on the temperature. Its all very technical. I'm not sure if mine will work because I didn't mix the tiny little drops when I added them to the tube.
Tiny Tiny Little PCR Tubes:

Tiny Tiny Little Clear Drops:

Thursday, March 10, 2016
DNA Extraction Number 2
This week in lab I performed another DNA extraction on my most recent web samples as well as the positive control web I got from ASU West. Earlier today I ran two tests to see how good my extraction was.
The first test I did is called a Nanodrop. This machine basically vaporizes a biological sample, shines a light through it, and using wave length absorbency can tell you the concentrations of biochemicals like proteins or nucleic acids or what have you. The concentrations of nucleic acids in my samples varied widely, which could have been a result of the varying amount of silk collected at each site. I cant really understand the rest of the data the Nanodrop gave me. There were graphs and stuff but it was incomprehensible to me. I'll have to learn about that later.
I also ran a gel today to see if there are any visible bands of DNA in my sample. The gel looked lovely. The first two lanes were an extraction process that Josh developed being done on known widow webs, the next three were my extraction process on unknown webs, the last visible band was extracted from a web built by a known widow with my extraction process, the next lane was a negative control and the last lane was empty. The four lanes I did my extraction process on had clear bands of genomic DNA. Even if the concentrations varied widely, at least I know the DNA is there.
The first test I did is called a Nanodrop. This machine basically vaporizes a biological sample, shines a light through it, and using wave length absorbency can tell you the concentrations of biochemicals like proteins or nucleic acids or what have you. The concentrations of nucleic acids in my samples varied widely, which could have been a result of the varying amount of silk collected at each site. I cant really understand the rest of the data the Nanodrop gave me. There were graphs and stuff but it was incomprehensible to me. I'll have to learn about that later.
#
|
Sample ID
|
User name
|
Date and Time
|
Nucleic Acid
|
Unit
|
A260 (Abs)
|
A280 (Abs)
|
260/280
|
260/230
|
Sample Type
|
Factor
|
1
|
J1
|
biology
|
3/10/2016 11:22 AM
|
31.8
|
ng/µl
|
0.635
|
0.399
|
1.59
|
0.28
|
DNA
|
50.00
|
2
|
J2
|
biology
|
3/10/2016 11:37 AM
|
40.7
|
ng/µl
|
0.814
|
0.463
|
1.76
|
0.22
|
DNA
|
50.00
|
3
|
HW1
|
biology
|
3/10/2016 11:38 AM
|
4.8
|
ng/µl
|
0.095
|
0.078
|
1.22
|
-1.24
|
DNA
|
50.00
|
4
|
RSW1
|
biology
|
3/10/2016 11:39 AM
|
13.5
|
ng/µl
|
0.269
|
0.229
|
1.18
|
2.28
|
DNA
|
50.00
|
5
|
FSE1
|
biology
|
3/10/2016 11:40 AM
|
7.7
|
ng/µl
|
0.155
|
0.095
|
1.62
|
1.84
|
DNA
|
50.00
|
6
|
+C1
|
biology
|
3/10/2016 11:42 AM
|
19.6
|
ng/µl
|
0.392
|
0.301
|
1.30
|
0.68
|
DNA
|
50.00
|
7
|
-C1
|
biology
|
3/10/2016 11:43 AM
|
1.5
|
ng/µl
|
0.029
|
0.017
|
1.76
|
-0.13
|
DNA
|
50.00
|
I also ran a gel today to see if there are any visible bands of DNA in my sample. The gel looked lovely. The first two lanes were an extraction process that Josh developed being done on known widow webs, the next three were my extraction process on unknown webs, the last visible band was extracted from a web built by a known widow with my extraction process, the next lane was a negative control and the last lane was empty. The four lanes I did my extraction process on had clear bands of genomic DNA. Even if the concentrations varied widely, at least I know the DNA is there.
Thursday, March 3, 2016
Spring 2016 Web Collection
This week in lab I collected spider webs for an extraction next week. I went around campus and collected the webs of a few spiders that looked like they could possibly be black widows. I also went to ASU West and collected some webs from their lab spiders. I'm hoping the webs spun in captivity will have less contaminates than webs found in the environment so I can use them as a positive control. The DNA I extract from unknown webs on campus will be compared to the DNA extracted from the lab webs as well as a molecular weight ladder.
Thursday, February 25, 2016
Spring 2016 Running the Gel
This week in lab I took the DNA I extracted last week and ran a gel electrophoresis gel with it. The protocol I followed was used to extract DNA from black widow webs, and I wanted to be sure that the extraction process worked on other types of spider webs as well, so I ran this gel to prove that I actually had extracted DNA from a variety of webs. This is what the gel looked like:
And here is the same gel with molecular weights and lane detection added in by the computer the fancy new gel imaging system can do:
So the thing is I put DNA in all but the last three wells but only two lanes are showing up on the lane detector. This means that I probably don't have DNA in those wells and the reasons for this are: not enough supernatant was moved from the extraction tube, or I left the DNA at room temperature too long and the DNA degraded, or I didn't use DNase free micro-centrifuge tubes and the DNA degraded, or I left the gels in the light too long and the syber green dye degraded, or I pipetted the DNA and loading dye wrong and didn't get any in the actual wells, or any number of other things. There doesn't seem to be a strong correlation between the type of spider the web was collected from, or the age of the web, and whether or not DNA was detected. In lane four, or two on the lane detection image, there is a strong clear band at 120bp. This was a web that was collected almost as it was being spun, as the spider was still in the web and construction was not completed yet, which may explain why the band is so clear.
And here is the same gel with molecular weights and lane detection added in by the computer the fancy new gel imaging system can do:
So the thing is I put DNA in all but the last three wells but only two lanes are showing up on the lane detector. This means that I probably don't have DNA in those wells and the reasons for this are: not enough supernatant was moved from the extraction tube, or I left the DNA at room temperature too long and the DNA degraded, or I didn't use DNase free micro-centrifuge tubes and the DNA degraded, or I left the gels in the light too long and the syber green dye degraded, or I pipetted the DNA and loading dye wrong and didn't get any in the actual wells, or any number of other things. There doesn't seem to be a strong correlation between the type of spider the web was collected from, or the age of the web, and whether or not DNA was detected. In lane four, or two on the lane detection image, there is a strong clear band at 120bp. This was a web that was collected almost as it was being spun, as the spider was still in the web and construction was not completed yet, which may explain why the band is so clear.
Thursday, February 18, 2016
Spring 2016 First DNA Extraction
This week in lab I performed my first spider web DNA extraction and my second extraction ever!
Whenever you try something new you learn a few things. While doing extractions this week I learned: I should make sure my microcentrifuge tubes are free of DNA-degrading enzymes called DNases, there's a big difference between 1.5 mL and 2.0 mL when working with the tiny volumes used in DNA extractions, and that to keep DNases from being activated, the DNA must be on ice nearly constantly.
I am unsure if my extraction went well enough for me to prove I have DNA from webs just yet but I'll get it soon!

Whenever you try something new you learn a few things. While doing extractions this week I learned: I should make sure my microcentrifuge tubes are free of DNA-degrading enzymes called DNases, there's a big difference between 1.5 mL and 2.0 mL when working with the tiny volumes used in DNA extractions, and that to keep DNases from being activated, the DNA must be on ice nearly constantly.
I am unsure if my extraction went well enough for me to prove I have DNA from webs just yet but I'll get it soon!

Thursday, February 11, 2016
Spring 2016 Week 3
Another awesome week in the lab.
This week I diluted ammonium acetate to be used in as a protein precipitating solution. All DNA is surrounded by a vast array of proteins. There are proteins in the cell walls and cytoplasm as well as chromatin proteins actually in the strands of DNA itself, wrapping the DNA into tightly wound bundles. The proteins can contaminate DNA samples, making tests hard to decipher. So to get rid of proteins and other cellular junk ammonium acetate is added to the solution. This caused the proteins to solidify out of solution, leaving the DNA behind.
This week I also collected some spider web samples to test my extraction process out on. I don't want to use up sources of black widow DNA just to make sure I can get DNA from webs, so I went out and collected samples from other species I could find on campus. I collected cellar spider webs from inside the lab, some abandoned looking web that could possibly have been a widow's web and the web from an orb weaver spider that is found on the chemistry department building. The orb webs were a little yellow in color, which I thought was super cool. The possible widow web might have too many leaves in it to use, but all the samples I collected from the environment had at least one contaminate it it: human hair, cat hair, leaves, dandelion seeds etc. Corri says that most of the contaminates will be washed out while the DNA is being processed, and because the primers we're building should be specific to spider DNA, only spider DNA should be amplified during PCR. We'll see.
Here is a picture I took of an extremely patient spider whose web I collected.
This week I diluted ammonium acetate to be used in as a protein precipitating solution. All DNA is surrounded by a vast array of proteins. There are proteins in the cell walls and cytoplasm as well as chromatin proteins actually in the strands of DNA itself, wrapping the DNA into tightly wound bundles. The proteins can contaminate DNA samples, making tests hard to decipher. So to get rid of proteins and other cellular junk ammonium acetate is added to the solution. This caused the proteins to solidify out of solution, leaving the DNA behind.
This week I also collected some spider web samples to test my extraction process out on. I don't want to use up sources of black widow DNA just to make sure I can get DNA from webs, so I went out and collected samples from other species I could find on campus. I collected cellar spider webs from inside the lab, some abandoned looking web that could possibly have been a widow's web and the web from an orb weaver spider that is found on the chemistry department building. The orb webs were a little yellow in color, which I thought was super cool. The possible widow web might have too many leaves in it to use, but all the samples I collected from the environment had at least one contaminate it it: human hair, cat hair, leaves, dandelion seeds etc. Corri says that most of the contaminates will be washed out while the DNA is being processed, and because the primers we're building should be specific to spider DNA, only spider DNA should be amplified during PCR. We'll see.
Here is a picture I took of an extremely patient spider whose web I collected.
Friday, February 5, 2016
Spring 2016 Week 2
This week in lab I began to gather and prepare the chemicals I will use for my DNA extraction.
To remove DNA from a eukaryotic sample, the first step is cracking open the cell's various membranes to get to the nucleus, where the DNA is stored. Lysis buffers are solutions of chemicals that break down the cellular membranes of a sample and also cause proteins to precipitate out of solution. Lysis buffers are generally alkaline and contain detergents, chemicals with both hydrophobic and hydrophilic portions of the molecule. These detergents and the alkalinity of the solutions cause the cellular membranes to degrade by disrupting the hydrophobic membranes. Other chemicals can be added to the lysis buffer to regulate pH and to cause unwanted macro-molecules, such as proteins, to precipitate.
So this week in lab I made my lysis buffer. None of the commercially available buffers had the same concentrations of chemicals as the protocol I'm following. I had to make 10mM Tris, 10mM EDTA, 2% SDS pH 8, from more concentrated chemicals found in the lab. It was great to practice dilution again. Diluting chemicals is a precise method that I feel is not one of my strengths and thus something I must practice more.
Thursday, January 28, 2016
Spring 2016 Week 1
Hello Everyone! I hope we all had a long relaxing winter break. I know I've been dying to get back to school and start working on my newest lab project.
This semester I plan to extract DNA from spider webs, and compare that to spider DNA stored in genomic databases. Spiders are notoriously difficult to classify; the juveniles of many species look markedly different from the adults as well as having extreme sexual dimorphism. To show that any two spiders belong to the same species DNA analysis is paramount. However, traditional methods of extracting DNA requires finding, capturing and killing spiders, a difficult task that reduces the overall number of spiders in an area and puts researchers at risk of being bitten.
In addition to the variety found among individuals of the same species, many spiders are secretive, only hunting at night and hiding during the day. To accurately assess the diversity of spiders in an area all species must be included, a nearly impossible task with current DNA extracting methods.
Using spider webs instead of spider body parts as a source of DNA solves both of these problems. The spiders no longer need to be handled and killed, reducing the harm to the spider and danger of envenomation to the researcher. Also many spiders build one web at night leaving it in the morning. Researchers can gather these refuse webs, extract DNA from them and get a more accurate idea of the number and species of spider living in an area.
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0142503
This semester I plan to extract DNA from spider webs, and compare that to spider DNA stored in genomic databases. Spiders are notoriously difficult to classify; the juveniles of many species look markedly different from the adults as well as having extreme sexual dimorphism. To show that any two spiders belong to the same species DNA analysis is paramount. However, traditional methods of extracting DNA requires finding, capturing and killing spiders, a difficult task that reduces the overall number of spiders in an area and puts researchers at risk of being bitten.
In addition to the variety found among individuals of the same species, many spiders are secretive, only hunting at night and hiding during the day. To accurately assess the diversity of spiders in an area all species must be included, a nearly impossible task with current DNA extracting methods.
Using spider webs instead of spider body parts as a source of DNA solves both of these problems. The spiders no longer need to be handled and killed, reducing the harm to the spider and danger of envenomation to the researcher. Also many spiders build one web at night leaving it in the morning. Researchers can gather these refuse webs, extract DNA from them and get a more accurate idea of the number and species of spider living in an area.
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0142503
Subscribe to:
Posts (Atom)