This week in lab is going to be the last one I ever do. This is such a surreal feeling. It seems like last week I was so excited to be a part of the program that I could barely process what had happened. Now its two years later and I'm transferring to the University of Arizona in a few short weeks. Life travels at such break neck speeds, making it nearly impossible to really comprehend what is happening while it happens.
How do you wrap up two years? I want to say deep meaningful things but all I can think is "where am I going to find a lab of such welcoming, interesting people ever again?" I'm going to miss you all so much! I can really be myself with you guys.
S-STEM Scholar Bethany Farah's Blog
Wednesday, May 4, 2016
Estrella Mountain Poster 2nd place winner
I guess I forgot to blog last week. Here is what my poster looked like by the time I was done with it.
I won second place!
Wednesday, April 13, 2016
The Slightly-Better Managed Week
Hey all! This week in lab has barely started. I'm blogging early so I can spend more time procrastinating on my research paper rough draft and poster. Whew I cant believe its already that time of year again!
Last week I ran one more PCR on my DNA samples to see if I could get them to work. This time Matt fiddled with the protocol a little bit, making the temperature changes' duration longer. We were hoping to get some better results this time.
Instead the results we got were strange and conflicting. The universal primer Matt gave me amplified all four of my samples but the widow primers I used worked less then 50% of the time. The strangest result was that none of the widow spider primers amplified the known widow spider web DNA (called +C in this experiment). Unfortunately the sample size I'm using is too small and does not allow for any real statistical analysis. I'm not really sure where to go from here.
Last week I ran one more PCR on my DNA samples to see if I could get them to work. This time Matt fiddled with the protocol a little bit, making the temperature changes' duration longer. We were hoping to get some better results this time.
Instead the results we got were strange and conflicting. The universal primer Matt gave me amplified all four of my samples but the widow primers I used worked less then 50% of the time. The strangest result was that none of the widow spider primers amplified the known widow spider web DNA (called +C in this experiment). Unfortunately the sample size I'm using is too small and does not allow for any real statistical analysis. I'm not really sure where to go from here.
Thursday, April 7, 2016
The Mismanaged Week
This week has been long and unproductive, or at least it feels that way. I feel that I don't manage my time as well as I could and end up paying for it at the end of the semester. I always feel like I have plenty of time to do things, put stuff off til the last minuet, and am always rushing to get things done.
So this Tuesday, instead of going to lab, I went to my mom's house and filed my taxes. I owe but no matter. So now I'm rushing at the end of the week to get some lab hours in.
After another inconclusive PCR I've decided to change some of the steps in the PCR protocol and see if that helps. If I don't get clear banding this time I'll have to run another DNA extraction because I'm out of DNA from one of my samples.
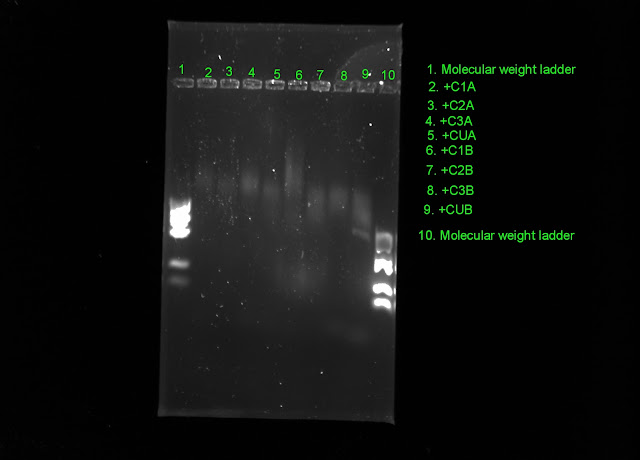
Here are my newest gels, see how the banding is not clear except HWUB and FSEUB. The A designation is the first PCR I did run through the thermocycler one more time to see if that increased visible bands. The B is another PCR I ran to see if the mixing of the sample was the problem.
So this Tuesday, instead of going to lab, I went to my mom's house and filed my taxes. I owe but no matter. So now I'm rushing at the end of the week to get some lab hours in.
After another inconclusive PCR I've decided to change some of the steps in the PCR protocol and see if that helps. If I don't get clear banding this time I'll have to run another DNA extraction because I'm out of DNA from one of my samples.
Here are my newest gels, see how the banding is not clear except HWUB and FSEUB. The A designation is the first PCR I did run through the thermocycler one more time to see if that increased visible bands. The B is another PCR I ran to see if the mixing of the sample was the problem.
Thursday, March 31, 2016
PCR Gels 1
This week in lab I took my final PCR product and ran it on a gel to see if the Polymerase Chain Reaction cut my DNA into the correct size of small fragments. The type of gel used for PCR products is exactly the same as the one used for genomic DNA, as well as the dye used to make DNA glow and the machine used to visualize the DNA. So in theory, if I had strong bands of genomic DNA after extraction, and I use the same gels and dye, as long as the PCR worked correctly, I should get pretty strong, clear bands of PCR product.
Unfortunately when I ran my PCR gels they did not come out as beautiful as my genomic DNA bands did. The PCR product was very difficult to visualize using the UV machine, and even where bands could be seen they were not of the right length for the primers I choose. I feel this might have been because I didn't mix my samples enough before putting them into the PCR machine. I am running another trial on my PCR to see if that was the case. If I still have weak bands I will have to trouble shoot the problem again.
Unfortunately when I ran my PCR gels they did not come out as beautiful as my genomic DNA bands did. The PCR product was very difficult to visualize using the UV machine, and even where bands could be seen they were not of the right length for the primers I choose. I feel this might have been because I didn't mix my samples enough before putting them into the PCR machine. I am running another trial on my PCR to see if that was the case. If I still have weak bands I will have to trouble shoot the problem again.
Friday, March 25, 2016
Dilution Fail
This week in lab I ran a Polyermerase Chain Reaction for the first time. I wanted to be able to have the PCR done on Thursday so I could run a gel on Friday but, alas, I did not account for the time it would take to prepare my chemicals and was not able to complete the experiment. All day Thursday I diluted my stock solutions of primers and my DNA samples to have consistent concentrations of each in my tiny test tubes.
To be honest I'm a little annoyed that I didn't know to get that stuff diluted on Tuesday when I had so little to do in the lab. From now on I need to take more initiative and find out exactly what I need to do myself, not rely on the lab instructor when they say I didn't have any prep work.
Anyway today I ran my PCR. One basically mixed together very small quantities of clear liquids into baby sized PCR tubes and then puts them into a machine that does all the work. The PCR machine cycles between various temperatures causing the clear liquids to do different steps of a reaction depending on the temperature. Its all very technical. I'm not sure if mine will work because I didn't mix the tiny little drops when I added them to the tube.
Tiny Tiny Little PCR Tubes:

Tiny Tiny Little Clear Drops:

To be honest I'm a little annoyed that I didn't know to get that stuff diluted on Tuesday when I had so little to do in the lab. From now on I need to take more initiative and find out exactly what I need to do myself, not rely on the lab instructor when they say I didn't have any prep work.
Anyway today I ran my PCR. One basically mixed together very small quantities of clear liquids into baby sized PCR tubes and then puts them into a machine that does all the work. The PCR machine cycles between various temperatures causing the clear liquids to do different steps of a reaction depending on the temperature. Its all very technical. I'm not sure if mine will work because I didn't mix the tiny little drops when I added them to the tube.
Tiny Tiny Little PCR Tubes:
Tiny Tiny Little Clear Drops:
Thursday, March 10, 2016
DNA Extraction Number 2
This week in lab I performed another DNA extraction on my most recent web samples as well as the positive control web I got from ASU West. Earlier today I ran two tests to see how good my extraction was.
The first test I did is called a Nanodrop. This machine basically vaporizes a biological sample, shines a light through it, and using wave length absorbency can tell you the concentrations of biochemicals like proteins or nucleic acids or what have you. The concentrations of nucleic acids in my samples varied widely, which could have been a result of the varying amount of silk collected at each site. I cant really understand the rest of the data the Nanodrop gave me. There were graphs and stuff but it was incomprehensible to me. I'll have to learn about that later.
I also ran a gel today to see if there are any visible bands of DNA in my sample. The gel looked lovely. The first two lanes were an extraction process that Josh developed being done on known widow webs, the next three were my extraction process on unknown webs, the last visible band was extracted from a web built by a known widow with my extraction process, the next lane was a negative control and the last lane was empty. The four lanes I did my extraction process on had clear bands of genomic DNA. Even if the concentrations varied widely, at least I know the DNA is there.
The first test I did is called a Nanodrop. This machine basically vaporizes a biological sample, shines a light through it, and using wave length absorbency can tell you the concentrations of biochemicals like proteins or nucleic acids or what have you. The concentrations of nucleic acids in my samples varied widely, which could have been a result of the varying amount of silk collected at each site. I cant really understand the rest of the data the Nanodrop gave me. There were graphs and stuff but it was incomprehensible to me. I'll have to learn about that later.
#
|
Sample ID
|
User name
|
Date and Time
|
Nucleic Acid
|
Unit
|
A260 (Abs)
|
A280 (Abs)
|
260/280
|
260/230
|
Sample Type
|
Factor
|
1
|
J1
|
biology
|
3/10/2016 11:22 AM
|
31.8
|
ng/µl
|
0.635
|
0.399
|
1.59
|
0.28
|
DNA
|
50.00
|
2
|
J2
|
biology
|
3/10/2016 11:37 AM
|
40.7
|
ng/µl
|
0.814
|
0.463
|
1.76
|
0.22
|
DNA
|
50.00
|
3
|
HW1
|
biology
|
3/10/2016 11:38 AM
|
4.8
|
ng/µl
|
0.095
|
0.078
|
1.22
|
-1.24
|
DNA
|
50.00
|
4
|
RSW1
|
biology
|
3/10/2016 11:39 AM
|
13.5
|
ng/µl
|
0.269
|
0.229
|
1.18
|
2.28
|
DNA
|
50.00
|
5
|
FSE1
|
biology
|
3/10/2016 11:40 AM
|
7.7
|
ng/µl
|
0.155
|
0.095
|
1.62
|
1.84
|
DNA
|
50.00
|
6
|
+C1
|
biology
|
3/10/2016 11:42 AM
|
19.6
|
ng/µl
|
0.392
|
0.301
|
1.30
|
0.68
|
DNA
|
50.00
|
7
|
-C1
|
biology
|
3/10/2016 11:43 AM
|
1.5
|
ng/µl
|
0.029
|
0.017
|
1.76
|
-0.13
|
DNA
|
50.00
|
I also ran a gel today to see if there are any visible bands of DNA in my sample. The gel looked lovely. The first two lanes were an extraction process that Josh developed being done on known widow webs, the next three were my extraction process on unknown webs, the last visible band was extracted from a web built by a known widow with my extraction process, the next lane was a negative control and the last lane was empty. The four lanes I did my extraction process on had clear bands of genomic DNA. Even if the concentrations varied widely, at least I know the DNA is there.
Subscribe to:
Comments (Atom)